Arbor Tracing
Microglia Tracing
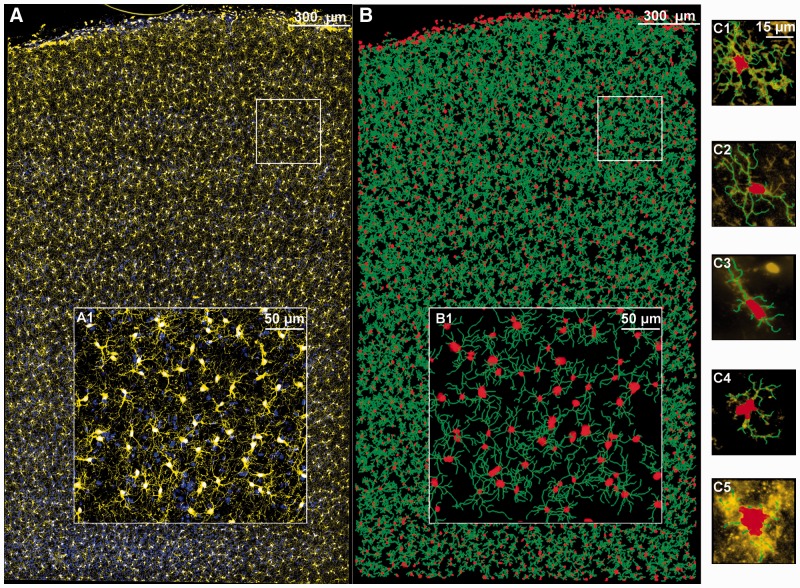
Motivation: The arbor morphologies of brain microglia are important indicators of cell activation. This paper fills the need for accurate, robust, adaptive, and scalable methods for reconstruct-ing 3-D microglial arbors & quantitatively mapping microglia acti-vation states over extended brain tissue regions.
Results: Thick rat brain sections (100-300m) were multiplex im-munolabeled for IBA1 and Hoechst, and imaged by step-and-repeat confocal microscopy with automated 3-D image mosa-icing, producing seamless images of extended brain regions (e.g., 5,903×9,874×229 voxels). An over-complete dictionary based model was learned for the image-specific local structure of microglial processes. The microglial arbors were reconstructed seamlessly using an automated and scalable algorithm that ex-ploits microglia-specific constraints. This method detected 80.1% and 92.8% more centered arbor points, and 53.5% and 55.5% fewer spurious points than existing vesselness and LoG based methods, respectively, and the traces were 13.1% and 15.5% more accurate based on the DIADEM metric. The arbor morpholo-gies were quantified using Scorcioni’s L-measure. Coifman’s har-monic co-clustering revealed four morphologically distinct clas-ses that concord with known microglia activation patterns. This enabled us to map spatial distributions of microglial activation and cell abundances.
Availability: Experimental protocols, sample datasets, scalable open-source multi-threaded software implementation (C++, MATLAB) in the electronic supplement, and website (www.farsight-toolkit.org).